CSC 533: Organization of Programming Languages
Spring 2008
HW6: More Scheme Programming
Place all of the function definitions for this assignment in a single file named
hw6.scm. Be sure to comment each function as to its behavior.
PART 1: Association Lists
In biology, the four bases that make up messenger RNA (mRNA) are adenine (A),
cytosine (C), guanine (G), and uracil (U). Within a cell, an mRNA sequence
is processed by a ribosome and translated into amino acids. Each grouping of 3
bases is known as a codon, and is translated to a particular amino acid.
For example, the codon GCU translates to Alanine.
The following Scheme association list maps each codon (represented by a triplet of
base symbols) to the abbreviation for the corresponding amino acid.
(define ACIDS
'( ((U U U) F) ((U U C) F) ((U U A) L) ((U U G) L)
((U C U) S) ((U C C) S) ((U C A) S) ((U C G) S)
((U A U) Y) ((U A C) Y) ((U A A) *) ((U A G) *)
((U G U) C) ((U G C) C) ((U G A) *) ((U G G) W)
((C U U) L) ((C U C) L) ((C U A) L) ((C U G) L)
((C C U) P) ((C C C) P) ((C C A) P) ((C C G) P)
((C A U) H) ((C A C) H) ((C A A) Q) ((C A G) Q)
((C G U) R) ((C G C) R) ((C G A) R) ((C G G) R)
((A U U) I) ((A U C) I) ((A U A) I) ((A U G) M)
((A C U) T) ((A C C) T) ((A C A) T) ((A C G) T)
((A A U) N) ((A A C) N) ((A A A) K) ((A A G) K)
((A G U) S) ((A G C) S) ((A G A) R) ((A G G) R)
((G U U) V) ((G U C) V) ((G U A) V) ((G U G) V)
((G C U) A) ((G C C) A) ((G C A) A) ((G C G) A)
((G A U) N) ((G A C) N) ((G A A) E) ((G A G) E)
((G G U) G) ((G G C) G) ((G G A) G) ((G G G) G) ))
- Define a function named translate that takes one input, a list
representing a codon, and returns the abbreviation for the corresponding
amino acid. For example, (translate '(G G A)) should evaluate to G.
- Define a function named translate-sequence that takes one input, an
arbitrarly long sequence of bases, and returns the corresponding list of amino acid
abbreviations. That is, the first symbol in the returned list should be the amino acid
corresponding to the first three bases, the second symbol should be the amino
acid corresponding to the next three bases, etc. If the length of the sequence
is not a multiple of three, any trailing bases should be ignored. For example,
(translate-sequence '(G G A U U C A C C C)) should evaluate to (G F T).
- When scientists extract mRNA samples from a cell, they invariably obtain
partial sequences. Since the start of the sequence may be unknown, it is unclear
exactly how the codons will be grouped in an actual translation. For example, in the sequence
GGAUUCACCC the codons could be grouped starting at the beginning
(yielding codons GGA UUC ACC), at the second base (yielding codons GAU UCA CCC),
or at the third base (yielding codons AUU CAC). In addition, it is possible that the
sequence has been read backwards, so the actual sequence to be translated should be
CCCACUUAGG.
Define a function named translate-all-possible that takes one input, an
arbitrarly long sequence of bases, and returns a list containing all six possible
translations of that sequence. Any extra bases at the beginning or end of the sequence
should be ignored in the translation. For example,
(translate-all-possible '(G G A U U C A C C C)) should evaluate to
((G F T) (N S P) (I H) (P T *) (P L R) (H L)).
PART 2: Structured Lists
In class, we discussed how structured lists could be used to represent
binary trees. For example, the following binary tree is represented by the
list to the right:
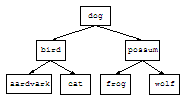
|
(define TREE1
'(dog
(bird (aardvark () ()) (cat () ()))
(possum (frog () ()) (wolf () ()))))
|
Several utility functions (empty?, root, left-subtree,
and right-subtree) were provided for you. Building upon these,
define the following Scheme functions that manipulate binary trees.
- Define a function named num-nodes that takes one input, a list
representing a binary tree, and returns the number of values stored in that tree.
For example, given the TREE1 structure above, (num-nodes TREE1) should
evaluate to 7.
- Define a function named leftmost that takes one input, a list
representing a binary tree, and returns the value that is stored leftmost in the
tree. For example, given the TREE1
structure above, (leftmost TREE1) should evaluate to aardvark.
Note: you may not assume that the input is a binary search tree, so the leftmost value
may not be the smallest value in the tree.
- Similarly, define a function named rightmost that takes one input, a list
representing a binary tree, and returns the value that is stored rightmost in the
tree. For example, given the TREE1
structure above, (rightmost TREE1) should evaluate to wolf.
Note: you may not assume that the input is a binary search tree, so the righmost value
may not be the largest value in the tree.
- Define a function named contains? that takes two inputs, a list
representing a binary tree and a value, and returns #t if the list
contains the value (otherwise #f). For example, given the TREE1
structure above, (contains? TREE1 'emu) should evaluate to #f
while (contains? TREE1 'frog) should evaluate to #t.
Note: you may not assume that the input is a binary search tree, so the
contains-bst? function from the lectures will not suffice here.
- Define a function named dupes? that takes one input, a list
representing a binary tree, and returns #t if the list
contains any duplicate values (otherwise #f). For example, given the TREE1
structure above, (dupes? TREE1) should evaluate to #f.

